By David McFadden
Bacteria have been dominating a kind of invisible arms race, constantly evolving tactics to evade the effects of antibiotics. It’s one of the world’s most pressing health threats, killing at least 700,000 people each year.
So-called “superbugs” are succeeding to such an alarming degree that one panel of international experts has forecasted that drug-resistant diseases could potentially cause 10 million deaths per year by 2050. The World Health Organization describes the issue as a “fundamental threat” to humanity.
Now, in a paper published in the journal Nature Communications, PhD candidate Michał Zieliński, Barry Sleno and Drs. Jaeok Park and Albert Berghuis from McGill University have reported new insights into how a class of antibiotics commonly used in both medicine and agriculture are rendered useless by resistant germs. Added to the existing pool of knowledge about antibiotic resistance, this latest contribution might one day help pave the way for next-generation antibiotics.
The findings can be thought of as the latest play in a very intricate puzzle, according to Dr. Berghuis, Chair of the Department of Biochemistry at McGill University’s Faculty of Medicine and Health Sciences and the paper’s senior author. “It’s one of those puzzle pieces that makes us understand more of how resistance works. And in many ways, every piece is important,” Berghuis said.
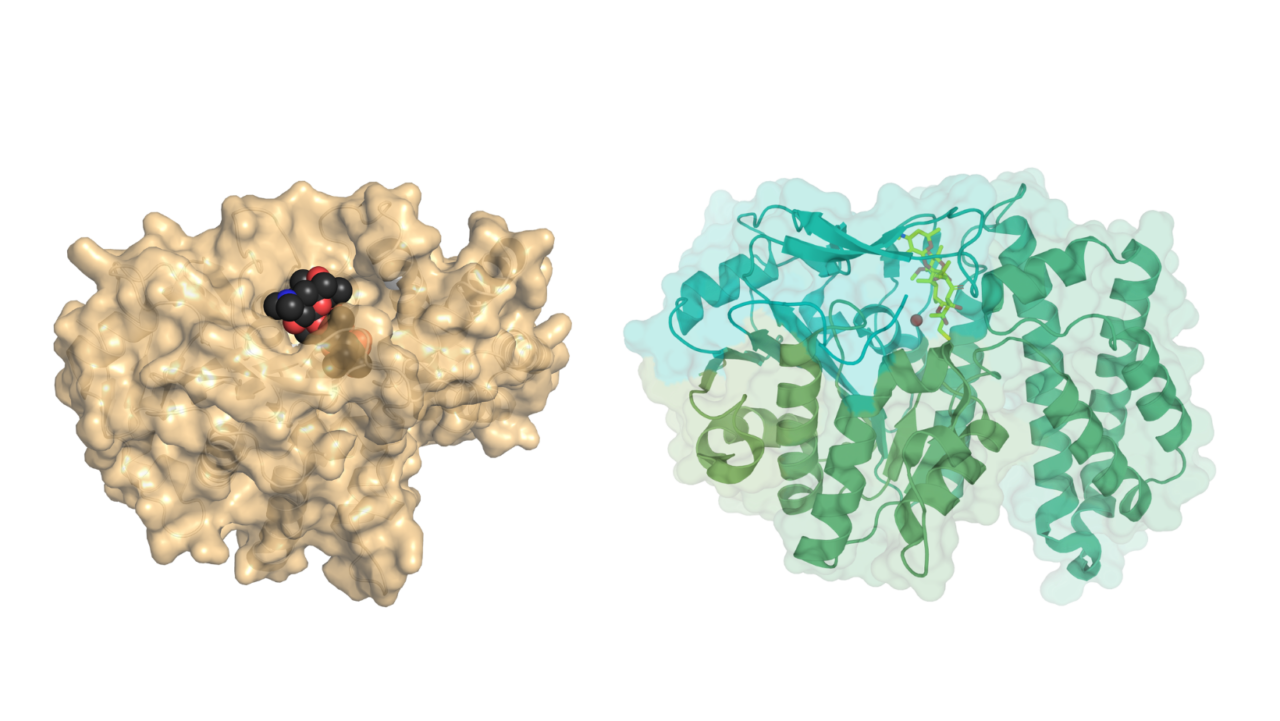
The new paper unveils structural details on how macrolides – widely-used antibiotics often prescribed to individuals allergic to penicillin – are stymied by bacterial resistance mechanisms. For the first-time, the three-dimensional structures of the “EreC” enzyme have been published. That’s significant because research into the under-the-radar Ere enzyme family has been relatively meager despite the outsized role it plays in macrolide resistance.
In a much-overlooked mechanism, the enzyme cleaves macrolides roughly akin to the way that dishwashing powder cuts through food waste on a plate. The most frequently identified Ere enzyme is “EreA,” which confers resistance to most clinically used macrolides. After analyzing their data, the McGill team discovered that “EreA” is nearly identical to the “EreC” enzyme, displaying more than 90 percent sequence identity. The McGill team also explored macrolide binding via molecular docking simulations and modeled the whole family of Ere enzymes. Persistence was key for the three-year research project. “Other people have tried, we were just the ones who got this to work,” Berghuis said.
The McGill team’s work relied on the Canadian Light Source synchrotron, a particle accelerator that provides researchers access to beamlines of ultra-intense light to run experiments seeking to understand the properties of materials down to their atomic detail. The researchers sent their samples packed in liquid nitrogen to Canada’s national synchrotron, located at the University of Saskatchewan.
Berghuis stressed that the battle against antibiotic resistance is a multipronged effort, dependent on the work of numerous scientists chipping away at the problem. It’s also dependent on common sense: Doctors cannot overprescribe antibiotics, patients must take them only as directed, and any use in crops and livestock must be managed with prudence.
Armed with their latest insights, Berghuis and his lab colleagues are now designing possible new macrolides on computers. With some luck and a whole lot more persistence, this research could potentially lead to new antibiotics you could get at your local pharmacy years down the line.
“Hopefully one of those many designs will indeed prove to be a much better antibiotic,” Berghuis said. “It’s not resulting in an antibiotic next week, but if we don’t know this stuff it’s going to become that much harder to overcome antibiotic resistance in the long-run.”
“Structural and functional insights into esterasemediated macrolide resistance,” by M. Zieliński, J. Park, B. Sleno and A. Berghuis was published in Nature Communications March 19, 2021. https://doi.org/10.1038/s41467-021-22016-3
