Source: Ludmer Centre for Neuroinformatics & Mental Health
Mathematical modeling is key to understanding the complex processes underpinning protein misfolding and neurological diseases.
Correctly folded, proteins play a central role in the structure, function, and regulation of the body’s tissues and organs, including the brain. Abnormal or misfolded proteins, in multiple studies, are increasingly associated with conditions such as Prion diseases, Alzheimer’s disease, Parkinson’s disease, Creutzfeldt–Jakob disease, amyotrophic lateral sclerosis (ALS), and several other degenerative disorders. However, the exact mechanisms and long-term impact on a patient’s clinical condition remain ill understood.
Dr Felix Carbonell at Biospective Inc and Ludmer Centre researchers Dr Yasser Iturria-Medina and Dr Alan Evans at the Montreal Neurological Institute hope to expedite research by providing an historical overview of the development of mathematical models for the aggregation and propagation of misfolded proteins in neurological diseases.
The authors also point out that neuroimaging (e.g., MRI, fMRI, PET) studies often fail to take full advantage of the tremendous capabilities of the ‘chemical kinetics modelling approach’ – mathematical models that describe the characteristics of a chemical reaction and how different experimental conditions can influence the speed of reaction.
Dr Carbonell said, “Our main goal was to connect the neuroimaging community with important studies of chemical kinetics modeling of misfolded proteins. Often under-acknowledged, we believe they constitute a solid framework for a better understanding of neurological evolution of diseases.”
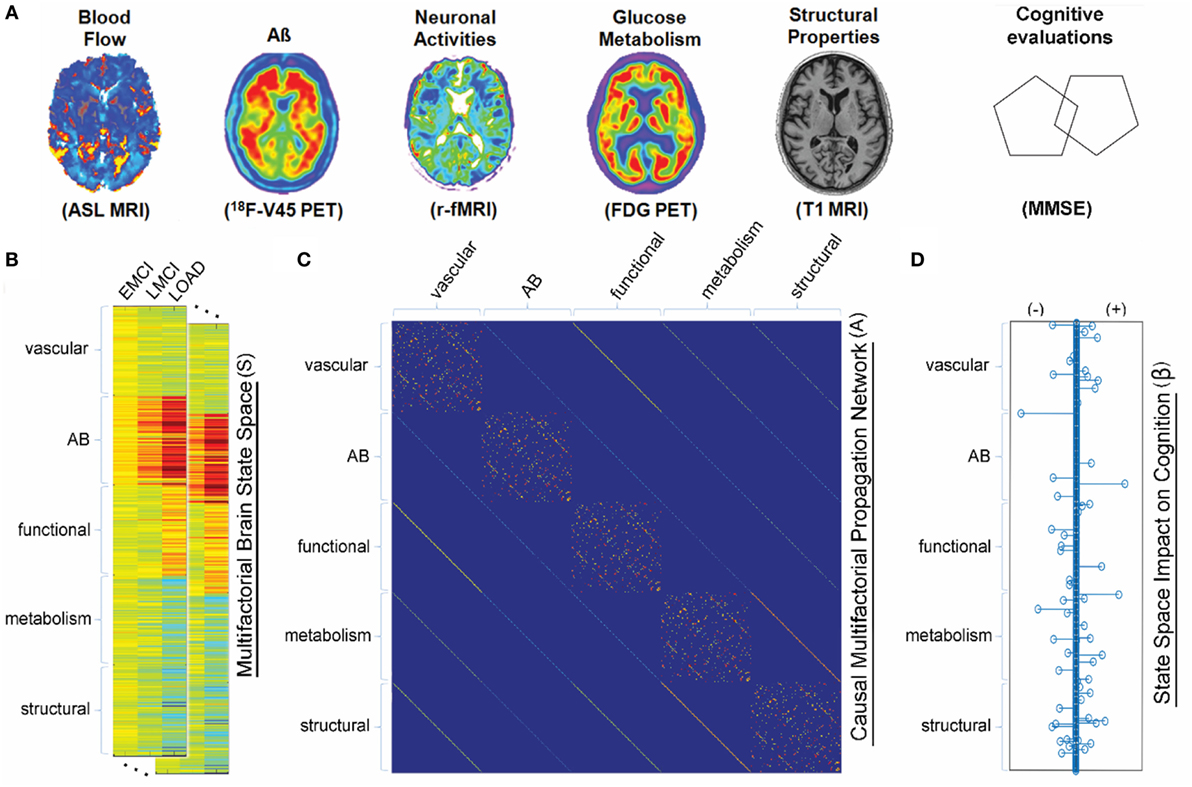
Our brains are the product of a multitude of ongoing molecular processes occurring at the microscopic level that cease only in death. To understand these processes and how they go awry to trigger neurological disorders, we need to study the living brain. Although yielding invaluable information, animal brains are not quite the same and human brain activity ceases in death, thus, animal proxy studies and cadaver-brain dissections have limitations. The closest we have gotten is when neuro-surgical patients agreed to have different brain centers stimulate by electrode probes and imaging technologies. Indeed, imaging (e.g., MRI, fMRI, PET), electrophysiological (e.g., EEG), genetic and epigenetic (e.g., DNA metalation) technologies, among others, are helping amass a growing tsunami of vital datasets. Yet, despite these advances, we have barely penetrated the brain’s “black box”.
Given the limitations in studying the living brain, how do we connect the wealth of datasets to better understand the underlying real-time processes? The answer is math, more precisely mathematical modelling of the fundamental formulas underpinning neurological processes.

An expert in multifactorial mathematical modeling, Dr Iturria-Medina said, “Our goal is to build models that describe complex neurological process from a multifactorial point of view, when and how neurodegenerative diseases develop and, ultimately, how these respond to specific pharmacological treatments.” Mathematical models underpin Dr Iturria-Medina’s work in precision medicine aimed at identifying individual brain signatures – personalized therapeutic fingerprints for the brain.
As highlighted in the publications, there is still a gap in our ability to properly model underlying microscopic processes of protein aggregation and their effects on the progression of neurological diseases. The researchers hope future pharmacokinetic modeling (maps the movement and impact of a drug into, though, and out of the body) PET-imaging studies of in-vivo protein binding might shed light on those unresolved issues and yield systematic drug discovery strategies under a broader, integrative perspective.
Dr Evans said, “Our goal is to increase our capacity to advance these models and to train new researchers in their application.”
February 6, 2018
